Recent Publications
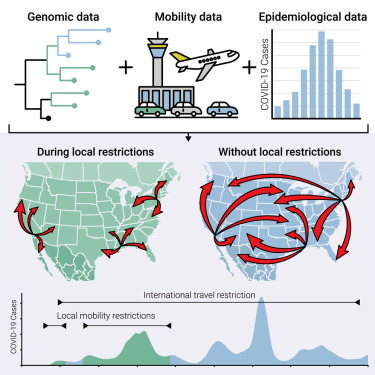
Elucidating human contact networks could help predict and prevent the transmission of SARS-CoV-2 and future pandemic threats. A new study from Scripps Research scientists and collaborators points to which public health protocols worked to mitigate the spread of COVID-19—and which ones didn’t. In the study, published online in Cell on December 14, 2023, the Scripps Research-led team of scientists investigated the efficacy…
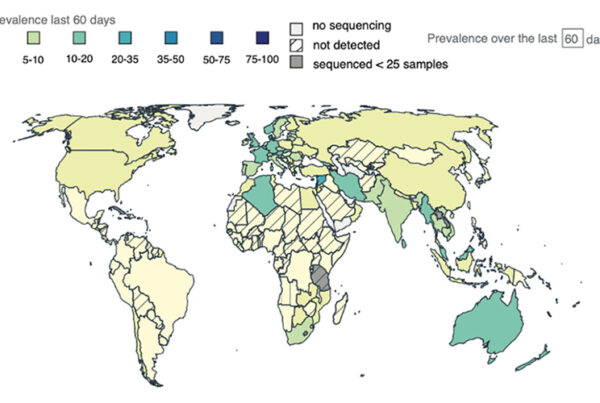
While COVID-19 may be transitioning from a “pandemic” to an “endemic” phase, it remains critically important to continue tracking the virus in real-time. In two new papers published in Nature Methods on Feb. 23, 2023, scientists at Scripps Research demonstrate the use of Outbreak.info as a standardized, searchable source of information on the COVID-19 virus and its many variants. Since the COVID-19 pandemic began,…
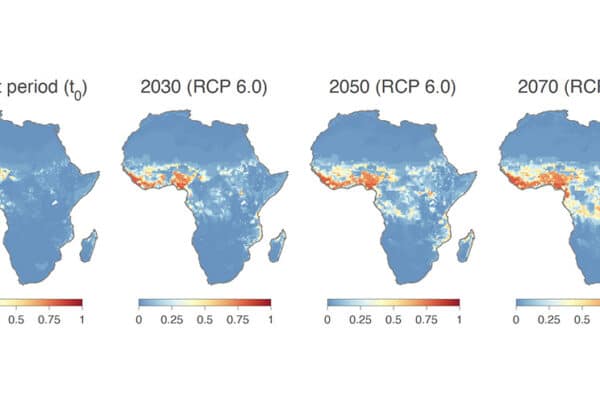
New analysis by scientists at Scripps Research and University of Brussels finds that climate change and other factors could soon make deadly Lassa fever a much bigger public health problem in Africa. In the study, which appeared on September 27, 2022, in Nature Communications, scientists analyzed decades of environmental data associated with Lassa virus outbreaks, revealing temperature, rainfall and the presence…